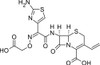
Cefixime Trihydrate is a broad-spectrum third generation cephalosporin targeting a wide range of Gram-positive and Gram-negative organisms. Cefixime Trihydrate is active against pathogens responsible for respiratory tract infections such as Haemophilus influenzae and Moraxella catarrhalis.
| Mechanism of Action | Like β-lactams, cephalosporins interfere with PBP (penicillin binding protein) activity involved in the final phase of peptidoglycan synthesis. PBP’s are enzymes which catalyze a pentaglycine crosslink between alanine and lysine residues providing additional strength to the cell wall. Without a pentaglycine crosslink, the integrity of the cell wall is severely compromised and ultimately leads to cell lysis and death. Resistance to cephalosporins is commonly due to cells containing plasmid encoded β-lactamases. Cefixime however, is resistant to many β-lactamases encoded by β-lactam resistant strains. |
| Spectrum | Cefixime Trihydrate is active against Gram-positive and Gram-negative bacteria |
| Microbiology Applications | Cefixime Trihydrate is commonly used in clinical in vitro microbiological antimicrobial susceptibility tests (panels, discs, and MIC strips) against Gram-positive and Gram-negative microbial isolates. Medical microbiologists use AST results to recommend antibiotic treatment options. Representative MIC values include:
Media SupplementsCefixime can be used as a selective agent in several types of isolation media: Sorbitol Macconkey Agar - Cefixime-Tellurite Supplement mTSB - VCC Selective Supplement CR-SMAC - Cefixime Supplement |
| Eukaryotic Cell Culture Applications |
Flavonoids from plants were able to alter the transport of Cefixime via PEPT1 in Caco-2 cells. Of the 33 flavonoids tested, quercetin, genistein, naringin, diosmin, acacetin, and chrysin increased Cefixime uptake by up to 60% via activating apical Na+/H+ exchange (Wenzel et al, 2001). Uptate of the anionic B-lactam Cefixime was studied using human Caco-2 human intestinal epitherlial cell line expressing intestinal peptide transporter (PEPT1). Researchers found that alterations of Ca2+ by Ca2+ channel blockers affect the pH of regulatory systems like apical Na+ and H+ exchange and alter the H+ gradient which is the driving force for uptake into epithelial cells. For example, at a surface pH of ≥ 6.25, the predominant fraction of Cefixime is charged, and thus shows only very low levels of interaction with PEPT1. The addition of the calcium blocker Nifedipine reduces the intracellular acidification induced by the influx of Cefixime and maintained a higher electrochemical proton gradient across the apical plasma membrane, allowing higher transport rates (Wenzel et al, 2002). |
| Molecular Formula | C16H15N5O7S2 • 3 H2O |
| Impurities | Chromatographic Purity: Individual Impurity: ≤1.0% Total Impurities: ≤2.0% |
| References |
Wenzel U, Kuntz S, Diestel S and Daniel H (2002) PEPT1-mediated Cefixime uptake into human intestinal epithelial cells is increased by Ca2+ channel blockers. Antimicrob. Agents. Chemother. 46(5):1375-1380 Wenzel U, Kuntz S and Daniel H (2001) Flavonoids with epidermal growth factor-receptor tyrosine kinase inhibitory activity stimulate PEPT1-mediated Cefixime uptake into human intestinal epithelial cells. J. Pharmacol. Exp. Ther. 299(1):351-357 PMID 11561098 |
| MIC | Bacteroides bivius| 1 - >32|| Bacteroides disiens| 1 - 16|| Bacteroides distasonis| 1 - >32|| Bacteroides fragilis| 1 - >128|| Bacteroides melaninogenicus| 16 - >32|| Bacteroides ovatus| 1 - >32|| Bacteroides thetaiotaomicron| 1 - >32|| Bacteroides vulgatus| 1 - >32|| Borrelia burgdorferi S.L.| 0.25 - 4|| Branhamella catarrhalis| 0.03 - 1|| Brucella abortus| 0.06 - 2|| Brucella melitensis| 0.06 - 2|| Brucella suis| 0.06 - 2|| Citrobacter amalonaticus| 0.5 - 1|| Citrobacter diversus| 0.03 - 0.25|| Citrobacter freundii| 0.12 - >32|| Clostridium bifermentans| >16 - >32|| Clostridium difficile| >16 - >64|| Clostridium histolyticum | 1 - 64|| Clostridium perfringens| 1 - 64|| Clostridium ramosum| >16 - >32|| Clostridium sordellii| 1 - 64|| Clostridium sporogenes| 1 - 64|| Clostridium spp.| >16 - >32|| Enterobacter aerogenes| 0.12 - >4|| Enterobacter agglomerans| ≤0.008 - >32|| Enterobacter cloacae| 0.12 - >64|| Enterobacter sakazakii| 0.015 - 0.5|| Enterobacteriaceae| 0.03 - 128|| Enterococci| 8 - 128|| Enterococcus faecalis| 4 ->128|| Enterococcus faecium| >64|| Enterococcus liquefaciens | >64|| Enterococcus spp.| >4|| Escherichia coli| 0.015 - 4|| Haemolytic streptococci| 0.03 - 0.5|| Haemophilus influenzae| ≤0.004 - >4|| Haemophilus parainfluenzae| 0.03 - 0.06|| Haemophilus spp.| 0.008 - 0.12|| Hafnia alvei| 1 - 4|| Helicobacter pylori| 0.03 - 8|| Hernia alvei| >0.5 - >32|| Klebsiella oxytoca| ≤0.125|| Klebsiella oxytoca (ceftazidime-resistant)| 0.015 - 2|| Klebsiella pneumonia| 0.03 - 8|| Klebsiella spp.| 0.004 - 4|| Kluyvera spp.| >0.5 - >32|| Listeria innocua| >64|| Listeria ivanovii| >64|| Listeria monocytogenes| >64|| Listeria seeligeri| >64|| Listeria welshimeri| >64|| Moraxella catarrhalis| 0.008 - 0.5|| Morganella morganii| 0.004 - >128|| Neisseria gonorrhoeae (penicillin-resistant)| 0.004 - 6|| Neisseria meningitidis| <0.06|| Neisseria spp.| 0.002 - 1|| Ochrobactrum anthropi (SLO74)| ≥128 - ?|| Peptococcus asaccharolyticus| >16 - >32|| Peptostreptococcus anaerobius| >16 - >32|| Pneumococci| 0.12 - 16|| Proteus mirabilis| ≤0.008 - 0.06|| Proteus vulgaris| ≤0.008 - 0.12|| Providencia alcalifaciens| >0.5 - >32|| Providencia rettgeri| ≤0.008 - 8|| Providencia spp.| 0.008 - 0.5|| Providencia stuartii| ≤0.008 - 32|| Pseudomonas aeruginosa| >4 - >64|| Pseudomonas cepacia| 32 - >64|| Pseudomonas flourescens| 8 - >64|| Pseudomonas putida| 8 - >64|| Pseudomonas stutzeri| 2 - >64|| Salmonella Agona| 0.03 - 0.13|| Salmonella Brandenburg| 0.03 - 0.13|| Salmonella enteritidis| 0.03 - 0.25|| Salmonella typhimurium| 0.03 - 0.13|| Serratia liquefaciens| 0.25 - >64|| Serratia marcescens| 0.12 - >64|| Serratia spp.| 0.12 - >128|| Shigella boydii| 1 - 4|| Shigella flexneri| 0.5 - >32|| Shigella sonnei| 0.5 - >32|| Shigella spp.| 0.12 - 1|| Staphylococci| 4 - 64|| Staphylococcus aureus| 2 - 128|| Staphylococcus cohnii| 8 - >64|| Staphylococcus epidermidis| ≤0.008 - >32|| Staphylococcus haemolyticus (methicillin-resistant)| 2 - >64|| Staphylococcus hominis| 2 - 8|| Staphylococcus pseudintermedius| ≤1 - 32|| Staphylococcus saprophyticus| 4 - 128|| Staphylococcus simulans| 4 - 16|| Staphylococcus spp. (coagulase-negative)| 1 - >4|| Staphylococcus warneri| 2 - 8|| Stenotrophomonas maltophilia| >32|| Streptococci| 0.06 - >8|| Streptococcus agalactiae| 0.25|| Streptococcus milleri (group)| 0.13 - 4|| Streptococcus mitior| 0.13 - 4|| Streptococcus pneumonia| 0.008 - 64|| Streptococcus pyogenes| 0.13 - 0.25|| Streptococcus spp. | ≤0.015 - 2|| Xanthomonas maltophilia| 4 - >64|| Yersinia enterocolitica| 0.06 - >8|| |