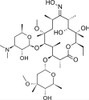
Clarithromycin related compound C, EvoPure (6-O-Methylerythromycin A (E)-9-Oxime) is a highly purified impurity found in Clarithromycin that can be used as a reference standard.
We also offer:
- Clarithromycin, USP (C033)
- Clarithromycin related compound D, EvoPure (C140)
- Clarithromycin related compound H, EvoPure (C143)
- Clarithromycin related compound I, EvoPure (C131)
- Clarithromycin related compound J, EvoPure (C132)
- Clarithromycin related compound K, EvoPure (C133)
- Clarithromycin related compound L, EvoPure (C134)
- Clarithromycin related compound M, EvoPure (C133)
- Clarithromycin related compound Z, EvoPure (E022)
| Application | Clarithromycin related compound C, EvoPure can be used as a reference standard. Reference standards are well characterized, highly pure compounds that can be used to help identify and/or quantify impurities in pharmaceutical compounds and antimicrobials. |
| Mechanism of Action | Macrolide antibiotics inhibit bacterial growth by targeting the 50S ribosomal subunit preventing peptide bond formation and translocation during protein synthesis. Resistance to Clarithromycin is commonly attributed to mutations in 50S rRNA preventing Clarithromycin binding allowing the cell to synthesize error-free proteins. Anti-cancer mechanisms include reduction of cytokines, inhibition of autophagy, and anti-angiogenesis. The compound can act on signal transduction pathways, transcription factors, drug pharmacokinetics, growth signals, and metastasis. These features can be exploited to make tumor cells more prone to apoptosis and reduce escape mechanisms. The mechanism used depends on the type of cancer. |
| Spectrum | Clarithromycin is a broad-spectrum antibiotic with bacteriostatic action on a wide range of Gram- positive and Gram-negative bacteria including anaerobes. It is also effective for Mycoplasma and Mycobacteria. |
| Microbiology Applications |
Clarithromycin is commonly used in clinical in vitro microbiological antimicrobial susceptibility tests (panels, discs, and MIC strips) against Gram-positive and Gram-negative bacteria. Medical microbiologists use AST results to recommend antibiotic treatment options. Representative MIC values include:
|
| Eukaryotic Cell Culture Applications | An in vitro study on the inhibitory effect of Clarithromycin on transporter-expressing HEK-293 cell lines or membrane vesicles with 13 drug transporters (OATP1B1, OATP1B3, OAT1, OAT3, OCT1, OCT2, MATE1, MATE2-K, P-gp, BCRP, MRP2, MRP3, and BSEP) were systematically assessed and results were that inhibition profiles were unique to the transporter (Vermeer et al, 2016). Macrolide antibiotics exert anti-inflammatory effects through inhibition of the production of proinflammatory cytokines. Nuclear factor kb (NF-kb) is an important transcription factor for genes that encode proinflammatory cytokines. In vitro studies with human monocytic leukemia cell line U-937, human T-cell leukemia (Jurkat), a pulmonary epithelial cell line A549, and peripheral blood mononuclear cells were conducted. Clarithromycin was found to suppress the production of proinflammatory cytokines via inhibition of NF-kb (Ichiyama T et al, 2001). Clarithromycin can have immunomodulatory activity on human lymphocyte function. The compound suppressed the synthesis of IL-1a, IL-1b, TNF-a, and GM-CSF in a concentration-dependent manner. This suggests Clarithromycin may modify the acute-phase inflammatory response by disturbing the cytokine cascade (Morikawa K et al, 1996). |
| Cancer Applications | Clarithromycin is involved in autophagy-lysosome pathway. It can inhibit autophagy in myeloma and myeloid leukaemia cells. It inhibits lysosomal function after fusion of the autophagosomes with the lysosomes. Thus, it could be a potential adjuvant where autophagy is used by the tumor as an escape mechanism (Nakamura et al, 2010). The combined treatment of clarithromycin with the proteasome inhibitor bortezomib enhances cytotoxicity in the breast cancer cell lines MDA-MB-231 and MDA-MB-468. A wild-type murine embryonic fibroblast (MEF) cell line also exhibited enhanced cytotoxicity (Komatsu et al, 2012). Direct antineplastic effects of CAM may depend on the tumor type. Researchers found a direct anti-tumor activity of CAM on lymphoma cells (Ochi et al, 2006) and it directly induced apoptosis in a murine B cell lymphoma cell line (Ohara et al, 2004). |
| Molecular Formula | C38H70N2O13 |
| References | Goldman RC, Zakula D, Flamm R, Beyer J, Capobianco J (1994) Tight binding of Clarithromycin, its 14-(R)-hydroxy metabolite, and erythromycin to Helicobacter pylori ribosomes. Antimicrob. Agents Chemother. 38(7):1496-500. PMID 7979278 Komatsu S et al (2012) Clarithromycin enhances bortezomib-induced cytotoxicity via endoplasmic reticulum stress-mediated CHOP (GADD153) induction and autophagy in breast cancer cells. Int. J. Oncol. 40(4):1029-1039 PMID 22200786 Tenson T, Lovmar M and Ehrenberg M (2003) The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330(5):1005-1014 PMID 12860123 Morikawa K, Watabe H, Araake M and Morikawa S (1996) Modulatory effect of antibiotics on cytokine production by human monocytes in vitro. Antimicrob. Agents. Chemother. 40(6):1366-1370 PMD 8726002 Nakamura M et al (2010) Clarithromycin attenuates autophagy in myeloma cells Int J Oncol 37:815–320 PMID 20811702 Niemi M, Neuvonen PJ, and Kivisto KT (2001) The cytochrome P4503A4 inhibitor clarithromycin increases the plasma concentrations and effects of repaglinide. 70(1):58-65 PMID 11452245 Ochi M et al (2006) Regression of primary low-grade mucosa-associated lymphoid tissue lymphoma of duodenum after long-term treatment with Clarithromycin. Scand J Gastroenterol 41:365–369 DOI: PMID: 16497629 Ohara T et al (2004) Antibiotics directly induce apoptosis in B cell lymphoma cells derived from BALB/c mice Anticancer Res. 24(6) pp 3723–30 Rose, WE et al (2015) Prevention of biofilm formation by methacrylate-based copolymer films loaded with rifampin, Clarithromycin, doxycycline alone or in combination. Pharm. Res. 32(1): 61-73 PMID 24934663 Ichiyama T et al (2001) Clarithromycin inhibits NF-κB activation in human peripheral blood mononuclear cells and pulmonary epithelial cells. Antimicrob. Agents. Chemother. 45 (1): 44-47 PMID 11120942 Van Nuffel AMT et al 2015 Repurposing drugs in oncology (ReDO)- Clarithromycin as an anti-cancer agent. Ecancer 9:513 Vermeer LMM, Isringhausen CD, Ogilvie BW and Buckley DB (2016) Evaluation of ketoconazole and its alternative clinical CYP3A4/5 inhibitors as inhibitors of drug transporters: The in vitro effects of ketoconazole, ritonavir, Clarithromycin, and itraconazole on 13 clinically-relevant drug transporters. Drug Metab. Disp. 44 (3) 453-459 PMID 26668209 |
| MIC | Bacillus subtilis (PCI219)| 0.1 - ?| 421| Bacteroides fragilis| 0.016 - >32| 948| Bacteroides fragilis| 0.03 - 2| 401| Bacteroides fragilis (NCTC 9343)| 0.25 - ?| 401| Bacteroides tectum| 0.125 - ?| 1460| Bacteroides ureolyticus| 0.125 - 4| 1460| Bilophila wadsworthia| 4 - 32| 1460| Borrelia afzelii (EB1)| 0.0312 - ?| 261| Borrelia afzelii (EB2)| 0.0078 - ?| 261| Borrelia afzelii (FAC-1)| 0.0078 - ?| 261| Borrelia afzelii (FEM1)| 0.0039 - ?| 261| Borrelia afzelii (VS461)| 0.0019 - ?| 261| Borrelia bissettii (25015)| 0.0078 - ?| 261| Borrelia burgdorferi| 0.008 - ?| 15| Borrelia burgdorferi (297)| 0.0019 - ?| 261| Borrelia burgdorferi (ATCC 35210 + B31)| 0.0039 - ?| 261| Borrelia burgdorferi (LW2)| 0.0039 - ?| 261| Borrelia burgdorferi (PKa-I)| 0.0156 - ?| 261| Borrelia burgdorferi (Z25)| 0.0019 - ?| 261| Borrelia garinii (A87SB)| 0.0019 - ?| 261| Borrelia garinii (JP2)| 0.0039 - ?| 261| Borrelia garinii (PSth)| 0.0312 - ?| 261| Borrelia garinii (PTrob)| 0.0039 - ?| 261| Borrelia garinii (ZQ1)| 0.0039 - ?| 261| Borrelia valaisiana (VS116)| 0.0039 - ?| 261| Brevibacterium spp.| ≤0.03 - >64| 185| Brevibacterium spp.| 0.03 - >128| 488| Burkholderia mallei (Pakistan)| ? - ?| 655| Campylobacter coli| 0.03 - 128| 277| Campylobacter helveticus (ATCC 51209)| 4 - ?| 421| Campylobacter jejuni| 0.03 - 128| 277| Campylobacter jejuni (ATCC 29428)| 0.25 - ?| 421| Campylobacter jejuni (ATCC 700819)| 1 - ?| 421| Chlamydia pneumonia| 0.015 - 0.06| 122| Chlamydia pneumonia| 0.015 - 0.06| 95| Chlamydia pneumonia| 0.015 - 0.12| 950| Chlamydia pneumonia| 0.015 - 0.125| 948| Chlamydia pneumonia| 0.015 - 0.125| 8| Chlamydia pneumonia (IOL-207)| 0.031 - ?| 963| Chlamydia pneumonia (KAjaani-6)| 0.031 - ?| 963| Chlamydia pneumonia (KKpn1)| 0.031 - ?| 963| Chlamydia pneumonia (KKpn15)| 0.016 - ?| 963| Chlamydia pneumonia (KKpn2)| 0.016 - ?| 963| Chlamydia pneumonia (KKpn6)| 0.016 - ?| 963| Chlamydia pneumonia (TW-183)| 0.015 - ?| 963| Chlamydia pneumonia (TW-183)| 0.016 - ?| 963| Chlamydia psittaci (Budgerigar)| 0.015 - ?| 963| Chlamydia psittaci (Izawa)| 0.015 - ?| 963| Chlamydia psittaci (MP)| 0.015 - ?| 963| Chlamydia trachomatis (C-HAR32)| ? - ?| 95| Chlamydia trachomatis (E-BOUR)| ? - ?| 95| Chlamydia trachomatis (F-IC-CAL3)| ? - ?| 95| Chlamydia trachomatis (J-UW-36)| ? - ?| 95| Chlamydia trachomatis (L2434)| ? - ?| 95| Chlamydophila pneumonia| 0.015 - 0.06| 1113| Clostridium clostridioforme| 0.125 - >32| 1460| Clostridium difficile| 0.125 - >32| 1460| Clostridium difficile| 0.5 - 128| 948| Clostridium innocuum| 0.25 - >32| 1460| Clostridium perfringens (GAI 5526)| 1 - ?| 397| Clostridium ramosum| 0.25 - >32| 1460| Corynebacterium afermentans| 0.03 - >128| 488| Corynebacterium amycolatum| 0.06 - >64| 185| Corynebacterium amycolatum| 1 - >128| 488| Corynebacterium jeikeium| 0.06 - >64| 185| Corynebacterium jeikeium| ≤0.015 - >128| 488| Corynebacterium minutissimum| ≤0.015 - 8| 488| Corynebacterium minutissimum| ≤0.03 - >64| 185| Corynebacterium pseudodiphtheriticum| ≤0.015 - 4| 488| Corynebacterium spp.| 0.008 - >64| 15| Corynebacterium striatum| 1 - >64| 185| Corynebacterium striatum| ≤0.015 - >128| 488| Corynebacterium urealyticum| ≤0.015 - 16| 488| Corynebacterium urealyticum| 0.125 - >64| 185| Coryneform (CDC group I4)| 0.03 - >128| 488| Edwardsiella hoshinae| 8 - 32| 1409| Edwardsiella ictaluri| 4 - >=128| 1409| Edwardsiella tarda| 8 - 32| 1409| Enterococci| 0.03 - 128| 401| Enterococci (erythromycin A-susceptible)| ≤0.06 - 4| 271| Enterococcus (erythromycin A-resistant)| 32 - >=512| 271| Enterococcus (penicillin-resistant)| 0.125 - >=512| 271| Enterococcus (penicillin-susceptible)| ≤0.006 - >=512| 271| Enterococcus (van(A))| ≥512 - ?| 271| Enterococcus faecalis (ATCC 29212)| 1.56 - ?| 380| Enterococcus faecalis (ATCC 29212)| 1.56 - ?| 421| Enterococcus faecium (ATCC 19434)| 1.56 - ?| 380| Enterococcus spp. (macrolide-resistant)| >128 - ?| 15| Enterococcus spp. (macrolide-susceptible)| 0.12 - 0.5| 15| Erysipelothrix rhusiopathiae| 0.06 - ?| 488| Escherichia coli| 32 - >128| 15| Escherichia coli (250 UC5)| 40 - ?| 58| Escherichia coli (K-12)| 6.25 - ?| 421| Fusobacterium gonidiaformans| <=0.015 - 32| 1460| Fusobacterium mortiferum| 4 - >32| 1460| Fusobacterium naviforme| <=0.015 - 32| 1460| Fusobacterium necrogenes| 4 - >32| 1460| Fusobacterium necrophorum| <=0.015 - 32| 1460| Fusobacterium nucleatum| <=0.015 - 32| 1460| Fusobacterium nucleatum subsp. animalis| <=0.015 - 32| 1460| Fusobacterium russii| 2 - >32| 1460| Fusobacterium ulcerans| 4 - >32| 1460| Fusobacterium varium| 32 - >32| 1460| Haemolytic streptococci| 0.015 - 16| 401| Haemophilus influenzae| 0.25 - 4| 1111| Haemophilus influenzae| 2 - 32| 15| Haemophilus influenzae| ? - ?| 403| Haemophilus influenzae| 0.008 - >=64| 950| Haemophilus influenzae| 0.03 - >128| 1113| Haemophilus influenzae| <0.05 - 128| 948| Haemophilus influenzae| ≤0.25 - >32| 500| Haemophilus influenzae| 0.25 - 32| 125| Haemophilus influenzae| <0.25 - >32| 85| Haemophilus influenzae| ≤0.5 - >64| 499| Haemophilus influenzae| 4 - 32| 1113| Haemophilus influenzae| 4 - 64| 769| Haemophilus influenzae| ? - ?| 1044| Haemophilus influenzae (351HT3)| 5 - ?| 58| Haemophilus influenzae (ampicillin-resistant + β-lactamase negative)| ≤0.5 - 32| 462| Haemophilus influenzae (ampicillin-resistant)| ? - ?| 58| Haemophilus influenzae (ampicillin-resistant)| 2 - >32| 265| Haemophilus influenzae (ampicillin-susceptible)| ? - ?| 58| Haemophilus influenzae (ampicillin-susceptible)| ≤0.25 - 32| 265| Haemophilus influenzae (ATCC 43095)| 6.25 - ?| 380| Haemophilus influenzae (ATCC 49247)| 4 - ?| 401| Haemophilus influenzae (ESBL + ampicillin-resistant)| 0.25 - >32| 18| Haemophilus influenzae (ESBL + ampicillin-resistant)| 0.25 - 32| 18| Haemophilus influenzae (ESBL + ampicillin-susceptible)| 0.25 - 32| 18| Haemophilus influenzae (ESBL)| <0.12 - >256| 45| Haemophilus influenzae (ESBL)| ≤0.5 - 32| 499| Haemophilus influenzae (ESBL)| 0.5 - 128| 45| Haemophilus influenzae (ESBL)| 2 - 32| 195| Haemophilus influenzae (HTM)| 4 - >32| 134| Haemophilus influenzae (MHF)| 4 - >32| 134| Haemophilus influenzae (NCTC 11931)| 8 - ?| 401| Haemophilus influenzae (non-ESBL)| 0.12 - >64| 195| Haemophilus influenzae (non-ESBL)| 0.5 - >64| 499| Haemophilus spp.| ? - ?| 403| Haemophilus spp.| 1 - 32| 401| Helicobacter bilis (ATCC 51630)| 8 - ?| 421| Helicobacter mustelae (ATCC 43772)| 1 - ?| 421| Helicobacter pullorum (ATCC 51864)| 2 - ?| 421| Helicobacter pylori| ≤0.007 - 64| 79| Helicobacter pylori| ≤0.004 - 0.03| 1010| Helicobacter pylori| 0.05 - 12.5| 662| Helicobacter pylori| 0.016 - 128| 105| Helicobacter pylori| 0.0156 - 64| 421| Helicobacter pylori| 0.05 - 0.1| 662| Helicobacter pylori| 0.063 - 0.25| 662| Helicobacter pylori (26695 + clarithromycin-susceptible)| 0.0156 - ?| 1228| Helicobacter pylori (26695 + clarithromycin-susceptible)| 31.25 - ?| 1228| Helicobacter pylori (ATCC 43504 + clarithromycin-resistant)| <0.8 - ?| 824| Helicobacter pylori (ATCC 43504 + pH 5.5)| 0.05 - ?| 778| Helicobacter pylori (ATCC 43504 + pH 7.0)| 0.012 - ?| 778| Helicobacter pylori (ATCC 43504)| 0.0078 - ?| 421| Helicobacter pylori (ATCC 43504)| 0.0156 - ?| 421| Helicobacter pylori (ATCC 43526 + pH 5.5)| 0.1 - ?| 778| Helicobacter pylori (ATCC 43526 + pH 7.0)| 0.012 - ?| 778| Helicobacter pylori (ATCC 43629 + pH 5.5)| 0.2 - ?| 778| Helicobacter pylori (ATCC 43629 + pH 7.0)| 0.05 - ?| 778| Helicobacter pylori (ATCC 43629)| 0.0313 - ?| 421| Helicobacter pylori (ATCC 49503 + pH 5.5)| 0.025 - ?| 778| Helicobacter pylori (ATCC 49503 + pH 7.0)| 0.006 - ?| 778| Helicobacter pylori (CPY 3281 + pH 5.4)| 0.5 - ?| 211| Helicobacter pylori (CPY 3281 + pH 7.4)| 0.031 - ?| 211| Helicobacter pylori (HCM 5940 + clarithromycin-resistant)| 9.8 - ?| 824| Helicobacter pylori (HCM 5972 + clarithromycin-resistant)| 7.8 - ?| 824| Helicobacter pylori (HCM 5994 + clarithromycin-resistant)| 6.3 - ?| 824| Helicobacter pylori (HCM 6000 + clarithromycin-resistant)| 12.5 - ?| 824| Helicobacter pylori (HCM 6002 + clarithromycin-resistant)| 4.7 - ?| 824| Helicobacter pylori (HCM 6026 + clarithromycin-resistant)| 37.5 - ?| 824| Helicobacter pylori (HCM 6034 + clarithromycin-resistant)| 6.3 - ?| 824| Helicobacter pylori (HCM 6072 + clarithromycin-resistant)| 25 - ?| 824| Helicobacter pylori (HOK-12 + metronidazole-resistant)| 0.025 - ?| 778| Helicobacter pylori (HOK-12 + metronidazole-resistant)| 0.025 - ?| 778| Helicobacter pylori (HOK-3 + clarithromycin-resistant)| 25 - ?| 778| Helicobacter pylori (KR 2098)| 0.0313 - ?| 421| Helicobacter pylori (macrolide-resistant)| 4 - 128| 15| Helicobacter pylori (macrolide-susceptible)| 0.008 - 0.06| 15| Helicobacter pylori (NCTC 11637 + pH 5.4)| 0.5 - ?| 211| Helicobacter pylori (NCTC 11637)| 0.5 - ?| 79| Helicobacter pylori (NCTC 11916)| 0.5 - ?| 79| Helicobacter pylori (pH 5.5)| 0.25 - ?| 658| Helicobacter pylori (pH 6.0)| 0.06 - ?| 658| Helicobacter pylori (pH 7.5)| 0.03 - ?| 658| Helicobacter pylori (SAP-3 + metronidazole-resistant)| 0.05 - ?| 778| Helicobacter pylori (SAP-9 + clarithromycin-resistant)| 6.25 - ?| 778| Helicobacter pylori (SHP 107)| 4 - ?| 79| Helicobacter pylori (SHP 133)| 2 - ?| 79| Helicobacter pylori (SWA-13 + metronidazole-resistant)| 0.05 - ?| 778| Helicobacter pylori (SWA-6 + clarithromycin-resistant)| 25 - ?| 778| Helicobacter pylori (SWA-7 + clarithromycin-resistant)| 25 - ?| 778| Helicobacter pylori (TH 1818)| 8 - ?| 421| Helicobacter pylori (TH 2095)| 0.0625 - ?| 421| Helicobacter pylori (TH 3391)| 0.0313 - ?| 421| Helicobacter pylori (TH 3392)| 0.0313 - ?| 421| Helicobacter pylori (TH 4165)| 0.0156 - ?| 421| Helicobacter pylori (TH 517)| 8 - ?| 421| Helicobacter pylori (TH 555)| 0.0313 - ?| 421| Helicobacter pylori (TH 582)| 0.0156 - ?| 421| Helicobacter pylori (TH 607)| 0.0625 - ?| 421| Helicobacter pylori (TH 627)| 0.0625 - ?| 421| Helicobacter pylori (TK 1003)| 0.0625 - ?| 421| Helicobacter pylori (TK 1025)| 0.0313 - ?| 421| Helicobacter pylori (TK 1027)| 0.0313 - ?| 421| Helicobacter pylori (TK 1030)| 0.0313 - ?| 421| Helicobacter pylori (TK 1042)| 0.0625 - ?| 421| Helicobacter pylori (TK 1047)| 2 - ?| 421| Helicobacter pylori (TK 1126)| 0.0313 - ?| 421| Helicobacter pylori (TK 1147)| 4 - ?| 421| Helicobacter pylori (TK 1307)| 0.0313 - ?| 421| Helicobacter pylori (TK 1308)| 0.0625 - ?| 421| Helicobacter pylori (TK 1310)| 0.0156 - ?| 421| Helicobacter pylori (TS 1131)| 4 - ?| 421| Helicobacter pylori (TS 119)| 0.0156 - ?| 421| Helicobacter pylori (TS 120)| 0.0156 - ?| 421| Helicobacter pylori (TS 1367)| 16 - ?| 421| Helicobacter pylori (TS 1407)| 32 - ?| 421| Helicobacter pylori (TS 1419)| 32 - ?| 421| Helicobacter pylori (TS 1445)| 32 - ?| 421| Helicobacter pylori (TS 1459)| 32 - ?| 421| Helicobacter pylori (TS 1556)| 32 - ?| 421| Helicobacter pylori (TS 1609)| 4 - ?| 421| Helicobacter pylori (TS 1611)| 8 - ?| 421| Helicobacter pylori (TS 1614)| 4 - ?| 421| Helicobacter pylori (TS 1664)| 8 - ?| 421| Helicobacter pylori (TS 1683)| 8 - ?| 421| Helicobacter pylori (TS 1711)| 16 - ?| 421| Helicobacter pylori (TS 1723)| 16 - ?| 421| Helicobacter pylori (TS 1729)| 8 - ?| 421| Helicobacter pylori (TS 1735)| 0.0156 - ?| 421| Helicobacter pylori (TS 1775)| 1 - ?| 421| Helicobacter pylori (TS 1826)| 8 - ?| 421| Helicobacter pylori (TS 1831)| 2 - ?| 421| Helicobacter pylori (TS 1832)| 4 - ?| 421| Helicobacter pylori (TS 1876)| 4 - ?| 421| Helicobacter pylori (TS 1887)| 4 - ?| 421| Helicobacter pylori (TS 1888)| 8 - ?| 421| Helicobacter pylori (TS 1889)| 0.0156 - ?| 421| Helicobacter pylori (TS 1890)| 4 - ?| 421| Helicobacter pylori (TS 1892)| 64 - ?| 421| Helicobacter pylori (TS 1893)| 32 - ?| 421| Helicobacter pylori (TS 1899)| 0.0313 - ?| 421| Helicobacter pylori (TS 251)| 0.0313 - ?| 421| Helicobacter pylori (TS 279)| 0.0156 - ?| 421| Klebsiella pneumonia (NCTC 9632)| 12.5 - ?| 421| Lactobacillus acidophilus (CRL 1251)| 10 - ?| 1268| Lactobacillus acidophilus (CRL 1266)| 1 - ?| 1268| Lactobacillus gasseri (CRL 1259)| >100 - ?| 1268| Lactobacillus johnsonii (CRL 1294)| 100 - ?| 1268| Lactobacillus paracasei (CRL 1289)| >100 - ?| 1268| Lactobacillus salivarius (CRL 1328)| 1 - ?| 1268| Legionella adelaidensis| 0.012 - ?| 32| Legionella anisa| 0.032 - ?| 32| Legionella birminghamiensis| 0.125 - ?| 32| Legionella bozemanii| 0.024 - ?| 32| Legionella brunensis| 0.064 - ?| 32| Legionella cherrii| 0.032 - ?| 32| Legionella cincinnatiensis| 0.032 - ?| 32| Legionella dumofii| 0.025 - ?| 32| Legionella erythra| 0.125 - ?| 32| Legionella fairfieldensis| 0.012 - ?| 32| Legionella feeleii| 0.05 - ?| 32| Legionella geestiana| 0.064 - ?| 32| Legionella gormanii| 0.039 - ?| 32| Legionella gratiana| 0.032 - ?| 32| Legionella hackeliae| 0.06 - ?| 32| Legionella israelensis| 0.032 - ?| 32| Legionella jamestowniensis| 0.125 - ?| 32| Legionella jordanis| 0.125 - ?| 32| Legionella lansingensis| 0.048 - ?| 32| Legionella londiniensis| 0.032 - ?| 32| Legionella longbeachae| 0.087 - ?| 32| Legionella maceachernii| 0.06 - ?| 32| Legionella micdadei| 0.053 - ?| 32| Legionella moravica| 0.032 - ?| 32| Legionella nautarum| 0.032 - ?| 32| Legionella oakridgensis| 0.06 - ?| 32| Legionella pneumophila| 0.001 - 0.004| 963| Legionella pneumophila| 0.002 - 0.03| 268| Legionella pneumophila| 0.004 - 0.125| 1113| Legionella pneumophila| 0.015 - 0.06| 948| Legionella pneumophila| 0.015 - 0.06| 950| Legionella pneumophila| 0.016 - 0.125| 32| Legionella quateirensis| 0.016 - ?| 32| Legionella quinlivanii| 0.032 - ?| 32| Legionella rubrilucens| 0.06 - ?| 32| Legionella sainthelensi| 0.046 - ?| 32| Legionella santicrucis| 0.046 - ?| 32| Legionella shakespearei| 0.064 - ?| 32| Legionella spiritensis| 0.032 - ?| 32| Legionella spp.| ≤0.004 - 0.03| 28| Legionella spp.| 0.06 - 0.12| 15| Legionella steigerwaltii| 0.016 - ?| 32| Legionella tucsonensis| 0.006 - ?| 32| Legionella wadsworthii| 0.032 - ?| 32| Legionella worsleiensis| 0.006 - ?| 32| Listeria monocytogenes| 0.06 - 0.125| 185| Listeria monocytogenes| 0.12 - ?| 15| Listeria spp.| 0.12 - ?| 488| Moraxella catarrhalis| 0.008 - 8| 1113| Moraxella catarrhalis| ≤0.03 - 0.25| 195| Moraxella catarrhalis| ≤0.03 - 0.25| 125| Moraxella catarrhalis| ≤0.03 - 0.25| 125| Moraxella catarrhalis| <0.03 - 8| 948| Moraxella catarrhalis| 0.008 - >=64| 950| Moraxella catarrhalis| 0.03 - 0.25| 86| Moraxella catarrhalis| ≤0.12 - 0.5| 499| Moraxella catarrhalis| ? - ?| 403| Moraxella catarrhalis| 0.12 - 0.25| 15| Moraxella catarrhalis| ≤0.25 - 1| 500| Moraxella catarrhalis| <0.25 - 16| 85| Moraxella catarrhalis| ≤0.25 - ?| 265| Moraxella catarrhalis (ESBL)| <0.015 - 0.5| 45| Moraxella catarrhalis (ESBL)| 0.03 - 0.12| 45| Moraxella catarrhalis (HTM)| 0.25 - ?| 134| Moraxella catarrhalis (MHF)| 0.25 - ?| 134| Morganella morganii (KONO)| >100 - ?| 421| Mycobacteria| ? - ?| 1084| Mycobacterium abscessus| 0.06 - 64| 291| Mycobacterium africanum (ATCC 25420 + pH 6.8)| 20 - ?| 61| Mycobacterium africanum (ATCC 25420 + pH 7.4)| 5 - ?| 61| Mycobacterium africanum (clinical isolate 901376 + pH 6.8)| 40 - ?| 61| Mycobacterium africanum (clinical isolate 901376 + pH 7.4)| 20 - ?| 61| Mycobacterium africanum (clinical isolate 92064 + pH 6.8)| 10 - ?| 61| Mycobacterium africanum (clinical isolate 920640 + pH 7.4)| 2.5 - ?| 61| Mycobacterium avium| ? - ?| 301| Mycobacterium avium| 0.6 - 2.5| 948| Mycobacterium avium (7H11 medium)| 6.25 - ?| 1080| Mycobacterium avium (7HSF medium)| 8 - ?| 1080| Mycobacterium avium (AIDS isolate HIV1423 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (AIDS isolate HIV1423 + pH 7.4)| 0.6 - ?| 61| Mycobacterium avium (AIDS isolate HIV733 + pH 6.8)| 1.25 - ?| 61| Mycobacterium avium (AIDS isolate HIV733 + pH 7.4)| 0.3 - ?| 61| Mycobacterium avium (AIDS isolate HIV804 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (AIDS isolate HIV804 + pH 7.4)| 1.25 - ?| 61| Mycobacterium avium (AIDS isolate HIV827 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (AIDS isolate HIV827 + pH 7.4)| 0.6 - ?| 61| Mycobacterium avium (ATCC 25291 + pH 6.8)| 0.6 - ?| 61| Mycobacterium avium (ATCC 25291 + pH 7.4)| 0.15 - ?| 61| Mycobacterium avium (clinical isolate 1110 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (clinical isolate 1110 + pH 7.4)| 0.6 - ?| 61| Mycobacterium avium (clinical isolate 1257 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (clinical isolate 1257 + pH 7.4)| 0.6 - ?| 61| Mycobacterium avium (clinical isolate 1295 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (clinical isolate 1295 + pH 7.4)| 1.25 - ?| 61| Mycobacterium avium (clinical isolate 711 + pH 6.8)| 1.25 - ?| 61| Mycobacterium avium (clinical isolate 711 + pH 7.4)| 0.3 - ?| 61| Mycobacterium avium (clinical isolate 969 + H 6.8)| 1.25 - ?| 61| Mycobacterium avium (clinical isolate 969 + pH 7.4)| 0.3 - ?| 61| Mycobacterium avium (macrolide-resistant)| >256 - ?| 15| Mycobacterium avium (macrolide-susceptible)| 0.25 - 32| 15| Mycobacterium avium-intracellulare complex| 0.125 - 64| 335| Mycobacterium avium-intracellulare complex| ? - ?| 1084| Mycobacterium bovis (ATCC 19210 + pH 6.8)| 10 - ?| 61| Mycobacterium bovis (ATCC 19210 + pH 7.4)| 2.5 - ?| 61| Mycobacterium bovis (BCG-Denmark + pH 6.8)| 0.15 - ?| 61| Mycobacterium bovis (BCG-Denmark + pH 7.4)| 0.07 - ?| 61| Mycobacterium bovis (BCG-Glaxo + pH 6.8)| 0.3 - ?| 61| Mycobacterium bovis (BCG-Glaxo + pH 7.4)| 0.15 - ?| 61| Mycobacterium bovis (BCG-Pasteur + pH 6.8)| 0.6 - ?| 61| Mycobacterium bovis (BCG-Pasteur + pH 7.4)| 0.3 - ?| 61| Mycobacterium avium (AIDS isolate HIV804 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (AIDS isolate HIV804 + pH 7.4)| 1.25 - ?| 61| Mycobacterium avium (AIDS isolate HIV827 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (AIDS isolate HIV827 + pH 7.4)| 0.6 - ?| 61| Mycobacterium avium (ATCC 25291 + pH 6.8)| 0.6 - ?| 61| Mycobacterium avium (ATCC 25291 + pH 7.4)| 0.15 - ?| 61| Mycobacterium avium (clinical isolate 1110 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (clinical isolate 1110 + pH 7.4)| 0.6 - ?| 61| Mycobacterium avium (clinical isolate 1257 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (clinical isolate 1257 + pH 7.4)| 0.6 - ?| 61| Mycobacterium avium (clinical isolate 1295 + pH 6.8)| 2.5 - ?| 61| Mycobacterium avium (clinical isolate 1295 + pH 7.4)| 1.25 - ?| 61| Mycobacterium avium (clinical isolate 711 + pH 6.8)| 1.25 - ?| 61| Mycobacterium avium (clinical isolate 711 + pH 7.4)| 0.3 - ?| 61| Mycobacterium avium (clinical isolate 969 + H 6.8)| 1.25 - ?| 61| Mycobacterium avium (clinical isolate 969 + pH 7.4)| 0.3 - ?| 61| Mycobacterium avium (macrolide-resistant)| >256 - ?| 15| Mycobacterium avium (macrolide-susceptible)| 0.25 - 32| 15| Mycobacterium avium-intracellulare complex| 0.125 - 64| 335| Mycobacterium avium-intracellulare complex| ? - ?| 1084| Mycobacterium bovis (ATCC 19210 + pH 6.8)| 10 - ?| 61| Mycobacterium bovis (ATCC 19210 + pH 7.4)| 2.5 - ?| 61| Mycobacterium bovis (BCG-Denmark + pH 6.8)| 0.15 - ?| 61| Mycobacterium bovis (BCG-Denmark + pH 7.4)| 0.07 - ?| 61| Mycobacterium bovis (BCG-Glaxo + pH 6.8)| 0.3 - ?| 61| Mycobacterium bovis (BCG-Glaxo + pH 7.4)| 0.15 - ?| 61| Mycobacterium bovis (BCG-Pasteur + pH 6.8)| 0.6 - ?| 61| Mycobacterium bovis (BCG-Pasteur + pH 7.4)| 0.3 - ?| 61| Mycobacterium simiae (AIDS isolate 96-005 + pH 6.8)| 20 - ?| 61| Mycobacterium simiae (AIDS isolate 96-005 + pH 7.4)| 5 - ?| 61| Mycobacterium simiae (AIDS isolate 96-012 + pH 6.8)| 20 - ?| 61| Mycobacterium simiae (AIDS isolate 96-012 + pH 7.4)| 5 - ?| 61| Mycobacterium simiae (ATCC 25275 + pH 6.8)| 20 - ?| 61| Mycobacterium simiae (ATCC 25275 + pH 7.4)| 10 - ?| 61| Mycobacterium smegmatis (A2058G)| >512 - ?| 1430| Mycobacterium tuberculosis (BACTEC 460 + H37Ra)| 1 - ?| 997| Mycobacterium tuberculosis (BACTEC 460 + H37Ragfp)| 0.5 - 2| 997| Mycobacterium tuberculosis (clinical isolate 900145 + pH 6.8)| 20 - ?| 61| Mycobacterium tuberculosis (clinical isolate 900145 + pH 7.4)| 5 - ?| 61| Mycobacterium tuberculosis (clinical isolate 900216 + pH 6.8)| 20 - ?| 61| Mycobacterium tuberculosis (clinical isolate 900216 + pH 7.4)| 5 - ?| 61| Mycobacterium tuberculosis (GFPMA H37Ragfp)| 0.5 - 3.2| 997| Mycobacterium tuberculosis (H37R + pH 6.8)| 20 - ?| 61| Mycobacterium tuberculosis (H37R + pH 7.4)| 5 - ?| 61| Mycobacterium tuberculosis (levofloxacin-resistant)| ? - ?| 301| Mycobacterium tuberculosis (levofloxacin-susceptible)| ? - ?| 301| Mycobacterium tuberculosis (multidrug-resistant)| ? - ?| 301| Mycobacterium tuberculosis (non-multidrug-resistant)| ? - ?| 301| Mycobacterium ulcerans (Angola-isolate + pH 6.6)| 0.25 - ?| 172| Mycobacterium ulcerans (Angola-isolate + pH 7.4)| 0.25 - ?| 172| Mycobacterium ulcerans (ATCC 19423 + 30℃ + ph 6.8)| 1.25 - ?| 61| Mycobacterium ulcerans (ATCC 19423 + 30℃ + pH 7.4)| 0.6 - ?| 61| Mycobacterium ulcerans (Australia-isolate + pH 6.6)| 0.5 - 4| 172| Mycobacterium ulcerans (Australia-isolate + pH 7.4)| <0.125 - 0.5| 172| Mycobacterium ulcerans (Benin-isolate + pH 6.6)| 0.125 - 1| 172| Mycobacterium ulcerans (Benin-isolate + pH 7.4)| <0.125 - 0.25| 172| Mycobacterium ulcerans (clinical isolate 94118 + 30℃ + ph 6.8)| 1.25 - ?| 61| Mycobacterium ulcerans (clinical isolate 94118 + 30℃ + pH 7.4)| 0.6 - ?| 61| Mycobacterium ulcerans (Co?te d’Ivoire-isolate + pH 6.6)| 0.25 - 0.5| 172| Mycobacterium ulcerans (Co?te d’Ivoire-isolate + pH 7.4)| 0.25 - 0.5| 172| Mycobacterium ulcerans (Democratic Republic of Congo-isolate + pH 6.6)| 0.125 - 1| 172| Mycobacterium ulcerans (Democratic Republic of Congo-isolate + pH 7.4)| <0.125 - 0.25| 172| Mycobacterium ulcerans (French Guiana-isolate + pH 6.6)| 0.5 - ?| 172| Mycobacterium ulcerans (French Guiana-isolate + pH 7.4)| 0.125 - ?| 172| Mycobacterium ulcerans (Ghana-isolate + pH 6.6)| 0.25 - ?| 172| Mycobacterium ulcerans (Ghana-isolate + pH 7.4)| 0.125 - 0.5| 172| Mycobacterium ulcerans (Malaysia-isolate + pH 6.6)| 2 - ?| 172| Mycobacterium ulcerans (Mexico-isolate + pH 6.6)| 0.125 - ?| 172| Mycobacterium ulcerans (Papua New Guinea-isolate + pH 6.6)| 0.25 - 0.5| 172| Mycobacterium ulcerans (Togo-isolate + pH 6.6)| 0.5 - ?| 172| Mycobacterium ulcerans (Togo-isolate + pH 7.4)| 0.125 - ?| 172| Mycoplasma fermentans| 8 - >32| 467| Mycoplasma fermentans| 16 - 32| 958| Mycoplasma fermentans| 16 - 32| 31| Mycoplasma genitalium| ≤0.015 - 0.03| 467| Mycoplasma genitalium| ≤0.015 - 0.06| 31| Mycoplasma genitalium| ≤0.015 - 0.06| 958| Mycoplasma hominis| >32 - ?| 467| Mycoplasma hominis| >64 - ?| 958| Mycoplasma hominis| >64 - ?| 31| Mycoplasma hyopneumoniae (F18)| 32 - ?| 761| Mycoplasma hyopneumoniae (F19)| >64 - ?| 761| Mycoplasma hyopneumoniae (F2)| 16 - ?| 761| Mycoplasma hyopneumoniae (F5)| 16 - ?| 761| Mycoplasma hyopneumoniae (F6)| 32 - ?| 761| Mycoplasma hyopneumoniae (J-strain)| 32 - ?| 761| Mycoplasma penetrans| 0.12 - 0.25| 467| Mycoplasma penetrans| 0.12 - ?| 31| Mycop |